|
Hello! I am a Research Assistant at NSU Optics Lab of North South University, where I work on computer vision and machine learning with a focus on medical imaging.
As a Reseach Assistant I work under supervision of Dr. M. Sohel Rahman and Dr. Mahdy Rahman Chowdhury
Email / CV / Google Scholar / Github / LinkedIn |

|
|
I'm interested in computer vision and machine learning with a focus on medical imaging (Microscopy, WSI, Histopathology, Radiology, Ultrasounds etc). In particular, my research interest lies in the following areas.
I am also actively involved in Quantum Computing research projects in NSU Optics and Quantum Mechanics Lab. |
|
|
|
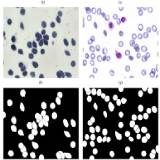
Authors: Deponker Sarker Depto, Shazidur Rahman, Md. Mekayel Hosen, Mst Shapna Akter, Tamanna Rahman Reme, Aimon Rahman, Hasib Zunair, M. Sohel Rahman, M.R.C. Mahdy Tissue and Cell, 2021 Paper / Dataset / Code / With the advent of deep learing algorithms in medial domain, there is a need for quality and large datasets. In this work, we introduced the largest microscopic blood cell segmentation dataset and benchmark different state-of-the-art algorithms on it. Our findings and contributions are particularly helpful for researchers working in deep learning with applications in medial domain. |
|
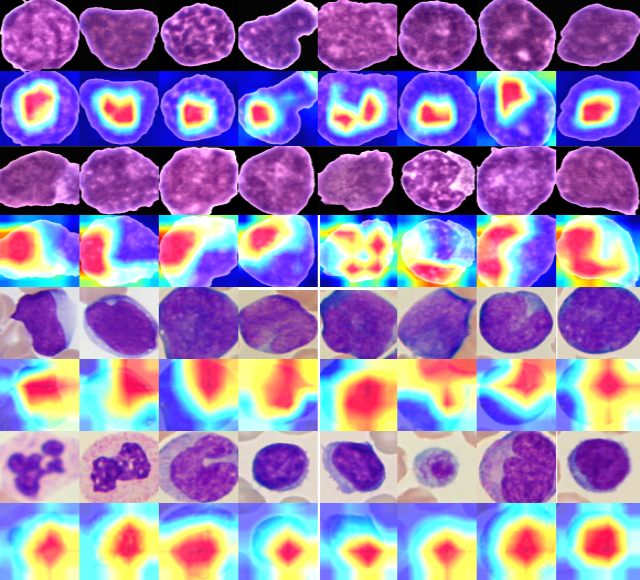
Deponker Sarker Depto, Md. Mashfiq Rizvee, Aimon Rahman, Hasib Zunair, M. Sohel Rahman, M.R.C. Mahdy Computers in Biology and Medicine, 2022 Paper / Code / Data In a plethora of imbalanced classification techniques, to decide on a specific one for imbalance handeling is an ambiguous task. With our effort, in this paper, we have performed extensive analysis on State-Of-The-Art imbalanced calssification techniques. We have presented emperical evidance that loss-based tecniques perform better than input-based and GAN-based techniques in high imbalance scenario. |
|
|
|
|